Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
RNA Profiling: More Help in Forensic Serology
Authors: Mr. Gaurav Kailas Tidke
DOI Link: https://doi.org/10.22214/ijraset.2024.64646
Certificate: View Certificate
Abstract
RNA profiling or, to be more specific mRNA profiling, is a very important method in the study of bio fluids. It is possible to obtain different type of body liquids from a crime scene. However, that requires more impromptu identification technique since the liquids undergo changes over time. In order to evaluate the different biomarkers present in the various bodily fluids which cannot be otherwise studied in usual procedures or to study the bodily fluids which are present in the backscattered toxic weapon, RNA profiling or writing is a non invasive technique. It can be very crucial in a case the distinction between menstrual blood and peripheral blood, for example, nasal mucosa and vaginal mucosa. For those body fluids, DNA profiling is the most common and well established when compared to RNA profiling. The results of those procedures seem to return a greater incidence of false positive results which are detrimental to the case in progress. In such cases, understanding RNA profiling assists in the development of a link with DNA profiling. cRNA is produced when multiple pre mRNAs are joined to form a circular structure—a step that is crucial in RNA profiling.
Introduction
I. INTRODUCTION
RNA or ribonucleic acid, a single chain polyribonucleotide controlled every important function in the early life this fact is not very acceptable. More clearly RNA was first genetic material. It is also not much clear how it helped as first biocatalyst1. Though it is not so sure still there are some enzymes – ribozymes are made up of RNA. Through some medical changes it formed into DNA. Now RNA acts as carrier of coded genetic information from DNA to cytoplasm to help in protein and enzyme synthesis.
RNA is single stranded polynucleotides but some places it shows partially double stranded due to folding or coiling of the single strand. There are 70-12000 ribonucleotides joined from start to end2. The backbone of RNA is made up of alternate residue of phosphate and ribose sugar. These phosphate combines with 5’ of its sugar and 3’ of next sugar and form a DNA like structure. There are four types of nitrogen bases present in RNA 1) adenine 2) guanine 3) cytosine 4) uracil. Nitrogen bases arrange as a complementary to respective DNA template. There are 3 types of RNA mainly help in every major classes. It also acts as a genetic material in some viruses.
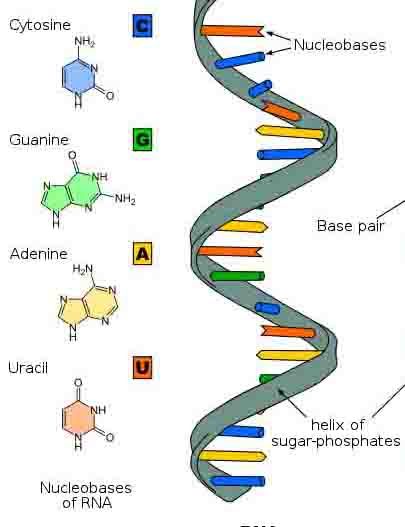
Source: https://i.pinimg.com/originals/d8/d4/bf/d8d4bf197be55e449fd4f4d436db3d16.jpg
Collected body fluids from the crime scene are major help to solve a particular crime. RNA was first mentioned in forensic literature by oehmichem et al in 1984 reporting on post mortal biosynthesis of DNA and RNA3. We can find RNA from dried blood stain, saliva mucosa, vaginal mucosa, buccal mucosa, semen. If there is very little amount of evidence, we still can apply PCR (polymerase chain reaction) to increase the amount of that specific biological fluid.
To work with body fluids, we have to be cautious about degradation of its. We should consider sensitive and stable bio markers to prevent the effect of degradation, for that we can consider microRNAs which are less prone to degradation due to their small size and stable structure.4 RNA profiling also helps to investigate postmortem time interval, age of the wound, identify the exact firearms used for murder from numerous numbers of fire arms in a gang fighting or robbery. But to get exact desirable results we should follow some precautions-1) how unbiasly we can execute the identification 2) distinguishable process adapt to extraction of RNA than DNA 3) consider physiological. Environmental factors. Through this article I am giving the idea of more use of RNAs in forensic investigation as it gives more accurate result than DNA profiling for body fluids and reduces false positive results. With the guidance of proper procedures, we can solve more cold cases. We will be able to give those cases a proper breakthrough and it will be easier to narrow down the suspects.
II. METHODS
Sample Collection ..Control samples ,each of which can be generated relatively easily from blood and different tissues obtained from a crime scene, or through the process of evidence collection, are being introduced. This control set includes blood, salina, vaginal mucosa, menstrual secretion, male and female subjects’ skin among four other subjects5. Taking fresh samples from the nasal mucosa would entail using both sides of the nostrils, and for diversity a larger number n will be a plus. From a total of 22 subjects, each individual sampled each one of the subject’s two nostrils, although in this case the subject was not coughing or ill. from seven of these, individuals were transfer paper for two deployment of 11 nosebleed slides from six. Also, 10 donors provided samples of sweat, tears and urine on the cotton swabs. There should be a distance to prevent skin-to-skin contact under semen aberrtraining is administered volunteers sex samples were obtained from four individuals. II. Two samples were taken from fertile males, and the other two were from post-vasectomized males. After that, we need to allow those willy swabs which have perforations and holes, to dry up and keep them in normal temperature as we are going to need them after sometime. Since a collection of 10 such samples was not successful, these samples were collected and kept in -20c and their wastes were also collected. 20 volunteers provided teaching penile swab sample using 4N6FLOQ swabs TM active drying mechanism by copnostic copandiagnostics7. All potential samples takers were informed and consent obtained before any sampling took place
|
ControlMarker name |
Tissue |
[primer] µM |
Forward primer (5’–3’) Reversed primer (5’–3’) |
Size (bp) |
Dye |
Reference |
|
CD93 |
Blood |
0.25 |
ACCAGTACAGTCCGACAC TTGCTAAGATTCCAGTCCAG |
151 |
NED™ |
8 |
|
HBB |
Blood |
0.035 |
GCACGTGGATCCTGAGAAC ATGGGCCAGCACACAGAC |
61 |
FAM™ |
8 |
|
HTN3 |
Saliva |
0.2 |
GCAAAGAGACATCATGGGTA GCCAGTCAAACCTCCATAATC |
134 |
VIC™ |
8 |
|
STATH |
Saliva/nasal mucosa |
0.3 |
TTTGCCTTCATCTTGGCTCT CCCATAACCGAATCTTCCAA |
93 |
FAM™ |
8 |
|
SEMG1 |
Seminal fluid |
0.8 |
GGAAGATGACAGTGATCGT CAACTGACACCTTGATATTGG |
91 |
FAM™ |
8 |
|
PRM1 |
Spermatozoa |
0.3 |
AGACAAAGAAGTCGCAGAC TACATCGCGGTCTGTACC |
146 |
NED™ |
8 |
|
CYP2B7P1 |
Vaginal mucosa |
0.8 |
AGTCTACCAGGGATATGGCATG CTATCAGACACTGAGCCTCGTCC |
141 |
VIC® |
9 |
|
MUC4 |
Vaginal mucosa |
0.8 |
CTGCTACAATCAAGGCCA AAGGGAAGTTCTAGGTTGAC |
88 |
FAM™ |
8 |
|
MMP7 |
Menstrual secretion |
0.8 |
GAACAGGCTCAGGACTATCTC TTAACATTCCAGTTATAGGTAGGCC |
107 |
VIC® |
8 |
|
MMP10 |
Menstrual secretion |
0.1 |
GCATCTTGCATTCCTTGTGCTGTTG GGTATTGCTGGGCAAGATCCTTGTT |
76 |
VIC® |
9 |
|
MMP11 |
Menstrual secretion |
0.4 |
CAACCGACAGAAGAGGTTCG GAACCGAAGGATCCTGTAGG |
71 |
NED™ |
9 |
Figure 1:(van den Berge & Sijen, 2017)
A. Presumptive Tests
after collecting all the specimen presumptive tests for every specimen are required individually. For semen the rapid stain identification series (RSID)is used and to observe the presence of spermatozoa, microscopic analysis has been also conducted. For saliva sample RSID has been also performed. TB testing for blood was performed by transferring biological material to a with water moistened filter paper, to which on drop of tetrabass solution in 10% acidic acid was added10 .one drop of barium per oxide solution was added next10 . The colour formation will observe according to criteria.
B. RNA Extraction
To extract RNA from biological sample we can proceed with commercial extraction kits. mirvanatm miRNA isolation kit11 . for RNA isolation we used the protocol described by lindenbergh et al12.DNase is used is to degrade. DNA in the process of RNA isolation. The RNA extracts were treated with DNase collected swabs were processed carefully. The swabs from excised from the nosebleed tissues were cut 1cm2 .if the amount of extracted DNA was below 1ng then RNA extracts from ethanol precipitated was prioritize in the reverse transcription.to separate non sperm fraction and sperm fraction ,the use of customized mild lysis is more effective. This mild lysis buffer is made up of phosphate buffered saline (PBS). which is composed with 1.6 mg proteinase k and 10 µM ribonucleoside vanadyl complex, these two inhibits various ribonucleases, in 50 µM of mild lysis buffer the swabs are incubated for 56 c for 20min.after using a QIAshredder column the lysate is separated from carrier material, during centrifugation at 11,000rpm.the nonsperm fraction and sperm fraction got separated. pellet of sperm fraction is washed using 400 µL PBS buffer with 10 µM ribonucleoside vanadyl complex and after that it is centrifuged at 13.200rpm for 5min.then the mirvana miRNA is isolation kit is used for binding lysis, then the supernatant of non-sperm fraction after centrifugation is treated with lysis buffer and then addition of homogenate additive is placed.
C. RNA Analysis
The methodology included CDNA synthesis followed by 19-plex reverse transcriptase PCR, product purification, and analysis implemented on a gene mapper. RNA markers identification enables the detection of these body fluids. Within this category exists a number of RNA markers for each fluid. Blood specifically uses markers HBB, CD93, AMICA1 while STATH, HTN3, KRT13 are for saliva and in some cases, SPRR2A is generalized for mucosal tissues. Semen or PRA1 other markers are added for Prm1 used for fertile men. MUC4 is used for vaginal mucosa markers and some others for menstrual fluid markers. Different types of markers perform useful in the classification of various body fluids. Nasal mucosa marker showed no cross reaction with BPIFA1 peripheral markers which can help in differentiating between blood from the periphery and the nose. Some markers don’t have positive results in the case of other body fluids in the distinction between the various types of body fluids.
III. RESULT
through the process of RNA typing, it helps us to differentiate between body fluids. but in case of DNA profiling, it provides the accurate results of the source of the individuals. RNA profiling or typing concludes the cell types present in evidentiary body fluids. The process of RNA typing is followed by clear guidelines. In case of vaginal mucosa due to presence of microbes and bacteria DNA profiling shows false positive result. But in case of examination of skin cells DNA profiling shows more false positive results for whenever skin contact with body fluids, though for detailed cell type information RNA typing is more helpful.
Conclusion
Since these chances are addressed, interrogators have particularly awed forensic serology with the introduction of RNA profiling pens, i.e. an approach that they have rated as informative. Some of these factors include: higher sensitivity and specificity, advancement in body fluids identification, origin of the tissue determined, distinction between individuals possible, among other factors. Moreover, RNA profiling offers an added advantage in forensic analysis of evidence as it can be used along with DNA analysis.Though progress has been made in the area, some concerns still persist such as the need to develop standards on the use of RNA, markers and techniques, degradation and contamination issues, and the establishment of effective working guidelines. More developments include: incorporating RNA profiling into the already existing forensic services and looking into moral and legal issues. RNA profiling has many applications in forensics, including: investigating a crime scene, analysis of stains, identification of different biological fluids, estimation of time after death, and forensic sciences - toxicology. Newer techniques like microRNA profiling and analysis of long non-coding RNAs are expected to lead to further improvements.The possibilities in forensic science will keep on increasing as further the study of RNA profiling evolves. Efforts to standardise and the work to integrate forensic scientists with researchers will propel the use of RNA profiling in criminal investigations as an indispensable technique.Generally, the high challenges in sensitivity, specificity as well as information value of RNA profiling technology makes it a worthy supplementary tool to forensic serology for more improved intensive criminal justice system and crime fighting capabilities.
References
[1] Bhatia, K. N., & Tyagi, M. P. (2014). Truemans: Elementary Biology. Vol. II, 27th edition, Trueman book company-New Delhi. [2] Oehmichen M, Zilles K. Postmortem DNA and RNA synthesis. Preliminary studies in human cadavers. Zeitschrift fur Rechtsmedizin. Journal of legal medicine. 1984;91(4):287. [3] Bhatia, K. N., & Tyagi, M. P. (2014). Truemans: Elementary Biology. Vol. II, 27th edition, Trueman book company-New Delhi. [4] van den Berge M, Bhoelai B, Harteveld J, Matai A, Sijen T. Advancing forensic RNA typing: on non-target secretions, a nasal mucosa marker, a differential co-extraction protocol and the sensitivity of DNA and RNA profiling. Forensic Science International: Genetics. 2016 Jan 1;20:119-29. [5] van den Berge M, Bhoelai B, Harteveld J, Matai A, Sijen T. Advancing forensic RNA typing: on non-target secretions, a nasal mucosa marker, a differential co-extraction protocol and the sensitivity of DNA and RNA profiling. Forensic Science International: Genetics. 2016 Jan 1;20:119-29. [6] van den Berge M, Bhoelai B, Harteveld J, Matai A, Sijen T. Advancing forensic RNA typing: on non-target secretions, a nasal mucosa marker, a differential co-extraction protocol and the sensitivity of DNA and RNA profiling. Forensic Science International: Genetics. 2016 Jan 1;20:119-29. [7] Alexander Lindenbergh, Mirjam de Pagter, Geeta Ramdayal, Mijke Visser, Dmitry Zubakov, Manfred Kayser, Titia Sijen,A multiplex (m)RNA-profiling system for the forensic identification of body fluids and contact traces,Forensic Science International: Genetics,Volume 6, Issue 5,2012,Pages 565-577, [8] M. van den Berge, A. Carracedo, I. Gomes, E.A.M. Graham, C. Haas, B. Hjort, P. Hoff-Olsen, O. Maroñas, B. Mevåg, N. Morling, H. Niederstätter, W. Parson, P.M. Schneider, D. Syndercombe Court, A. Vidaki, T. Sijen,A collaborative European exercise on mRNA-based body fluid/skin typing and interpretation of DNA and RNA results,Forensic Science International: Genetics,Volume 10,2014,Pages 40-48,ISSN 1872-4973 [9] Lindenbergh A, Maaskant P, Sijen T. Implementation of RNA profiling in forensic casework. Forensic Science International: Genetics. 2013 Jan 1;7(1):159-66 [10] Grabmüller M, Madea B, Courts C. Comparative evaluation of different extraction and quantification methods for forensic RNA analysis. Forensic Science International: Genetics. 2015 May 1;16:195-202. [11] Lindenbergh A, de Pagter M, Ramdayal G, Visser M, Zubakov D, Kayser M, Sijen T. A multiplex (m) RNA-profiling system for the forensic identification of body fluids and contact traces. Forensic Science International: Genetics. 2012 Sep 1;6(5):565-77. [12] .Zhang Y, Liu B, Shao C, Xu H, Xue A, Zhao Z, Shen Y, Tang Q, Xie J. Evaluation of the inclusion of circular RNAs in mRNA profiling in forensic body fluid identification. International journal of legal medicine. 2018 Jan 1;132(1):43-52. [13] Lux C, Schyma C, Madea B, Courts C. Identification of gunshots to the head by detection of RNA in backspatter primarily expressed in brain tissue. Forensic science international. 2014 Apr 1;237:62-9. [14] Benschop CC, Wiebosch DC, Kloosterman AD, Sijen T. Post-coital vaginal sampling with nylon flocked swabs improves DNA typing. Forensic science international: genetics. 2010 Feb 1;4(2):115-21.
Copyright
Copyright © 2024 Mr. Gaurav Kailas Tidke . This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET64646
Publish Date : 2024-10-17
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

